Charlie Harris (Charles)
PhD Student @ University of Cambridge
I’m finishing a PhD at Cambridge, where I develop generative and geometric deep-learning methods for structure-based drug discovery in the labs of Pietro Lio and Sir Tom Blundell.
Until recently, I served as Technical Lead in the UK Government’s Sovereign AI Unit. Working closely with the Prime Minister’s AI Adviser, Matt Clifford, I helped shape early thinking on AI for Science — particularly around the idea that targeted, high-quality datasets can be a practical lever for unlocking “AlphaFold-scale” advances.
In that role, I led the £8m seed investment to establish OpenBind, a consortium aimed at building large-scale protein–ligand datasets for AI-driven drug discovery, and supported a £5m DSIT expansion of the Encode AI for Science Fellowship. I also helped design the GPU compute allocation approach set out under Sovereign AI Compute, influencing how millions of GPU-hours were distributed, and contributed to the UK’s AI for Science Strategy.

Education
-
University of Cambridge
PhD in Computer Science Oct. 2021 - Present
-
Imperial College London
MSc in Bioinformatics and Theoretical Systems Biology Oct. 2020 - Sep. 2021
-
Imperial College London
BSc in Biochemisty Oct. 2017 - Jun. 2020
Experience
-
UK Government
Technical Lead (Opportunties), UK Sovereign AI Unit Mar. 2025 - Dec. 2025
-
IQ Capital
Venture Fellow June 2024 - Oct. 2024
-
BenevolentAI
Machine Learning Research Intern July 2022 - Oct. 2022
Honors & Awards
- UK-Italy Visiting Researcher Fellowship - Alan Turing Institute 2024
- Polaris Fellowship - Entrepreneur First 2024
- Cambridge Half Blue (for gliding) - Univesity of Cambridge 2023
- CCAIM PhD Studentship - University of Cambridge 2021
- Associateship - Royal College of Science 2020
- Gold - UK Chemistry Olympiad 2017
Media
News
Selected Publications (view all )
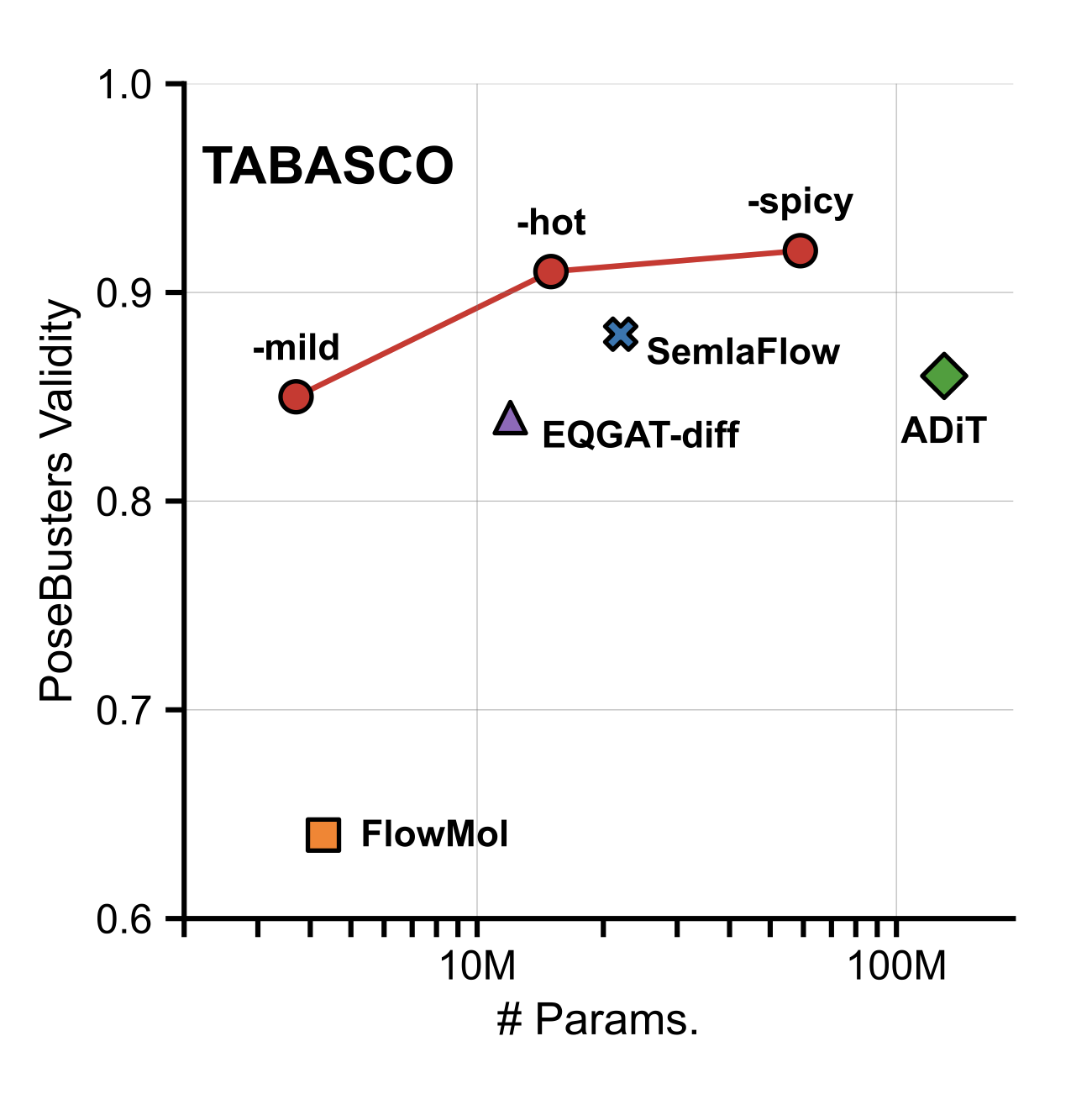
TABASCO: A Fast, Simplified Model for Molecular Generation with Improved Physical Quality
Carlos Vonessen*, Charles Harris*, Miruna Cretu*, Pietro Liò (* equal contribution)
Transactions on Machine Learning Research (TMLR)(Camera ready pending) 2026
We introduce TABASCO which re- laxes these assumptions: The model has a standard non-equivariant transformer architecture, treats atoms in a molecule as sequences and re- constructs bonds deterministically after genera- tion. The absence of equivariant layers and message passing allows us to significantly simplify the model architecture and scale data through- put. On the GEOM-Drugs benchmark TABASCO achieves state-of-the-art PoseBusters validity and delivers inference roughly 10× faster than the strongest baseline, while exhibiting emergent ro- tational equivariance despite symmetry not be- ing hard-coded.


DiffSBDD: Structure-based Drug Design with Equivariant Diffusion Models
Arne Schneuing*, Charles Harris*, Yuanqi Du*, Arian Jamasb, Ilia Igashov, Weitao Du, Tom Blundell, Pietro Lió, Carla Gomes, Max Welling, Michael Bronstein, Bruno Correia (* equal contribution)
Nature Computational Science 2024
DiffSBDD was one of the first equivariant diffusion models for structure-based drug design.

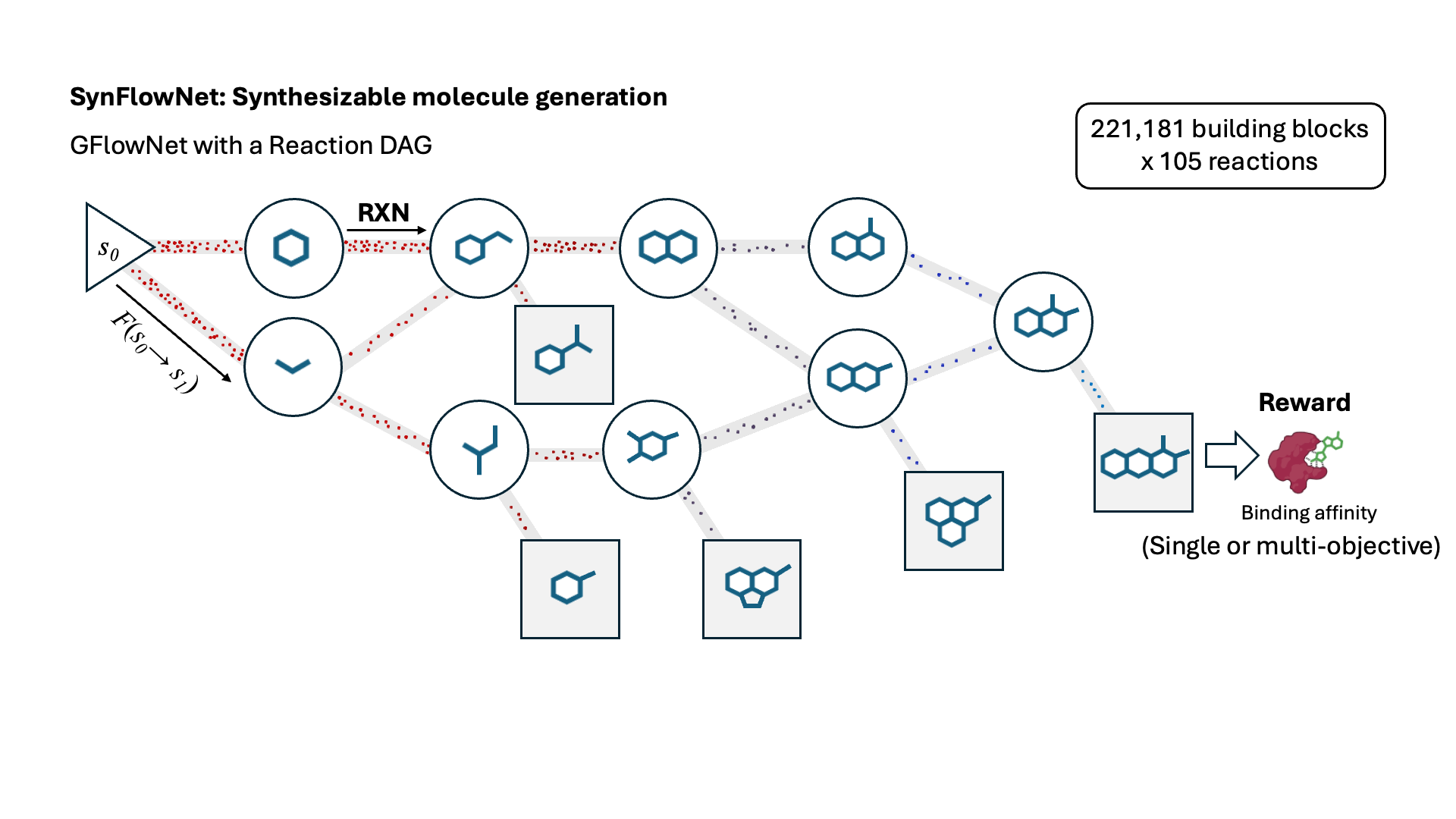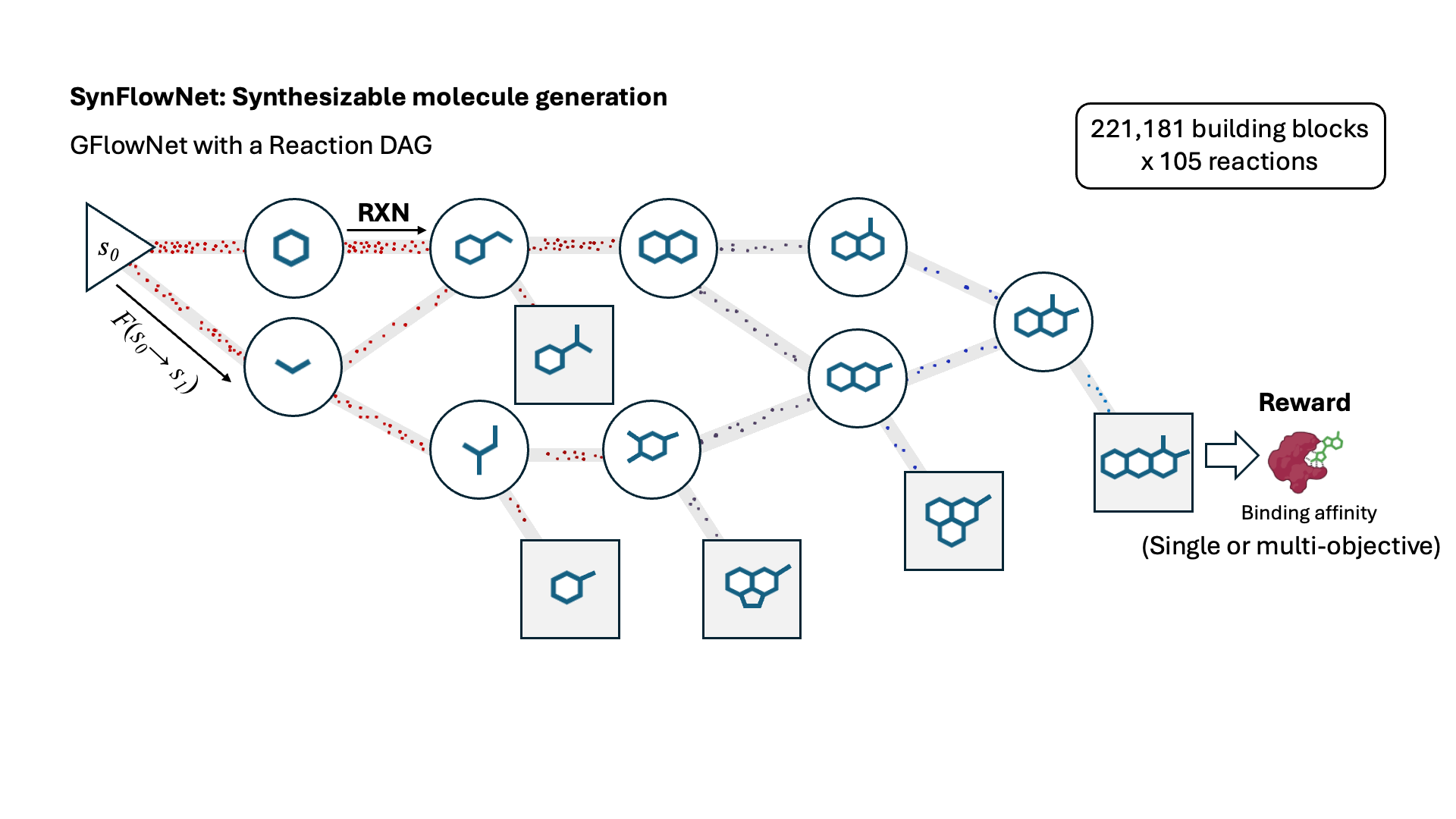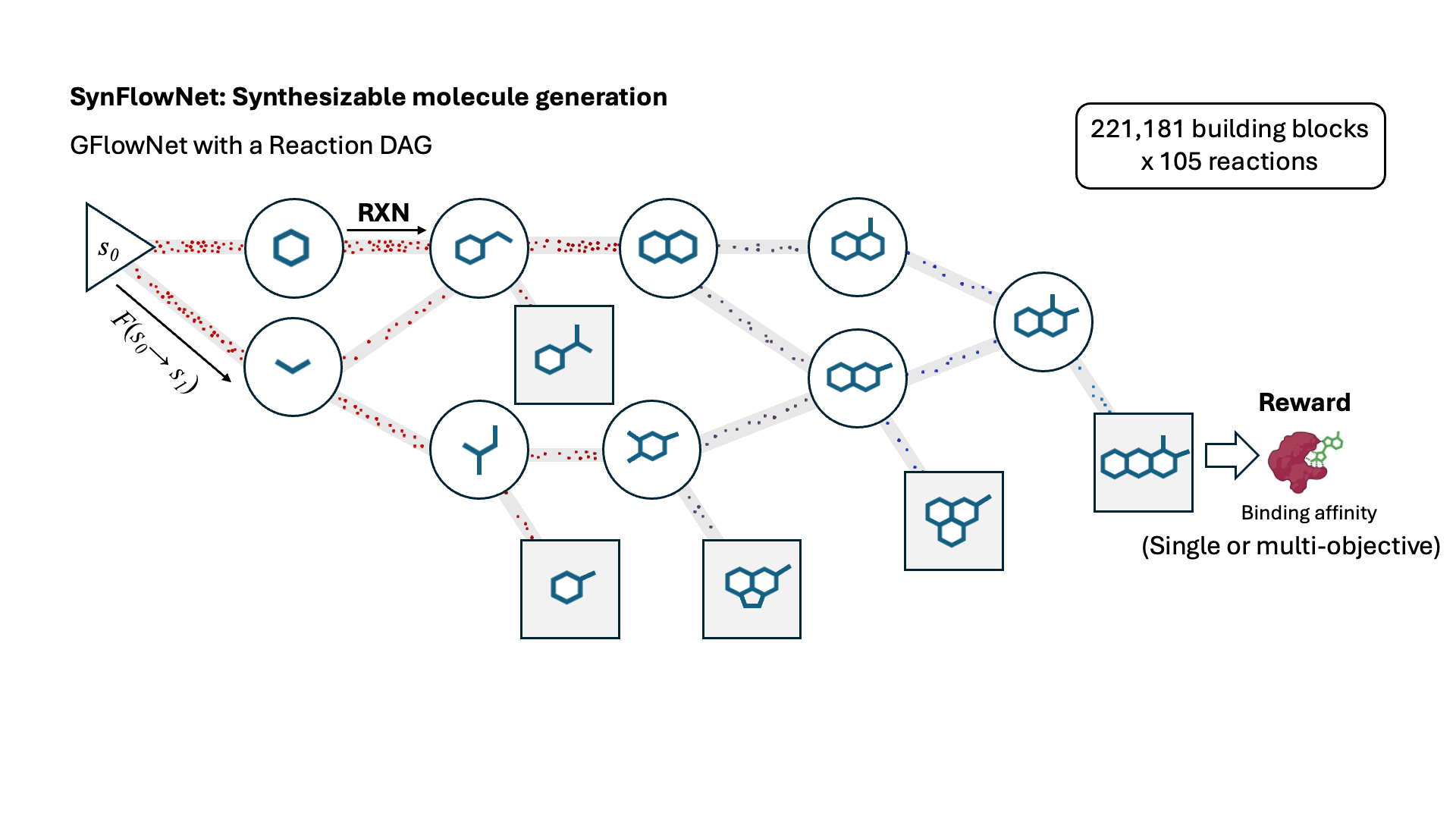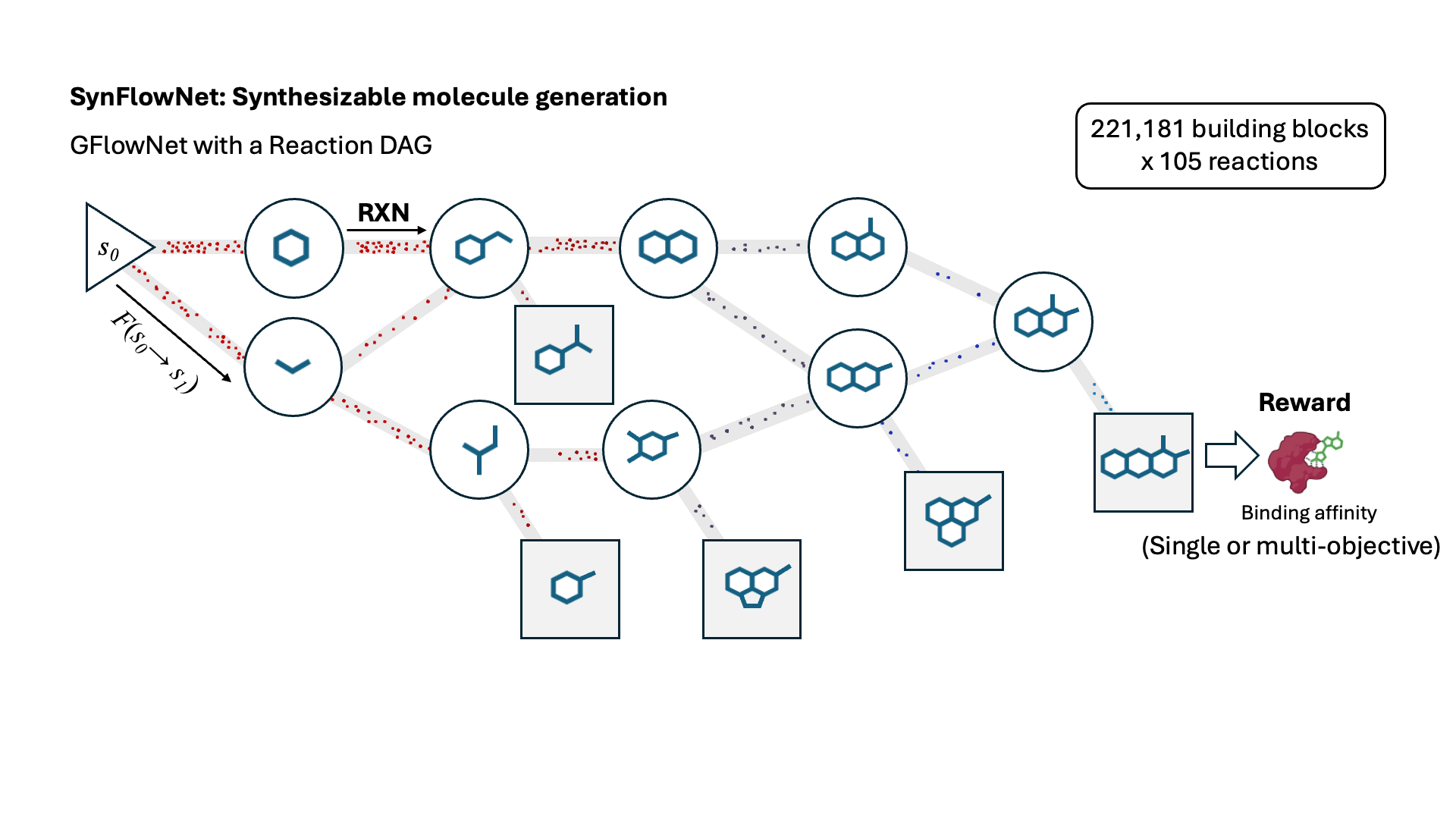
SynFlowNet: Design of Diverse and Novel Molecules with Synthesis Constraints
Miruna Cretu, Charles Harris, Ilia Igashov, Arne Schneuing, Marwin Segler, Bruno Correia, Julien Roy, Emmanuel Bengio, Pietro Liò
ICLR 2025 Spotlight
This work introduces SynFlowNet, a GFlowNet model whose action space uses chemically validated reactions and reactants to sequentially build new molecules. We evaluate our approach using synthetic accessibility scores and an independent retrosynthesis tool. SynFlowNet consistently samples synthetically feasible molecules, while still being able to find diverse and high-utility candidates.


PoseCheck: Generative Models for 3D Structure-based Drug Design Produce Unrealistic Poses
Charles Harris, Kieran Didi, Arian Jamasb, Chaitanya Joshi, Simon Mathis, Pietro Liò, Tom Blundell
MLSB Workshop @ NeurIPS 2023 Spotlight
This work introduced PoseCheck, an extensive analysis of multiple state-of-the-art methods and find that generated molecules have significantly more physical violations and fewer key interactions compared to baselines, calling into question the implicit assumption that providing rich 3D structure information improves molecule complementarity. We make recommendations for future research tackling identified failure modes and hope our benchmark will serve as a springboard for future SBDD generative modelling work to have a real-world impact.
