2025
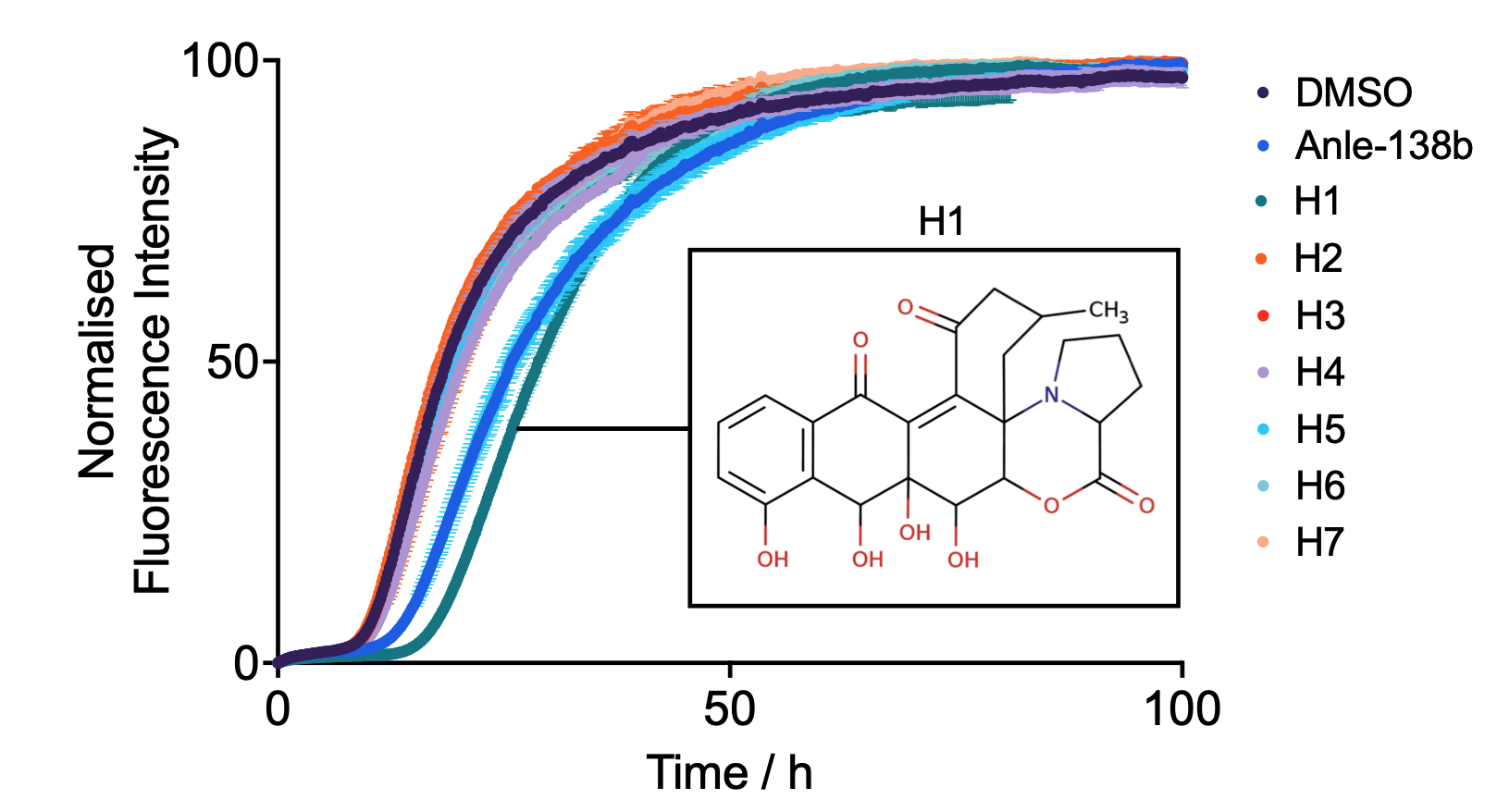
Structure-based Generation of a Secondary Nucleation Inhibitor in α-Synuclein Aggregation Using a Conditional Diffusion Model
Hao Zhang, Robert I. Horne, Faidon Brotzakis, Charles Harris, Pietro Liò, Michele Vendruscolo
Under review 2025
A conditional diffusion model is used to discover a potent inhibitor of secondary nucleation in α-synuclein aggregation.

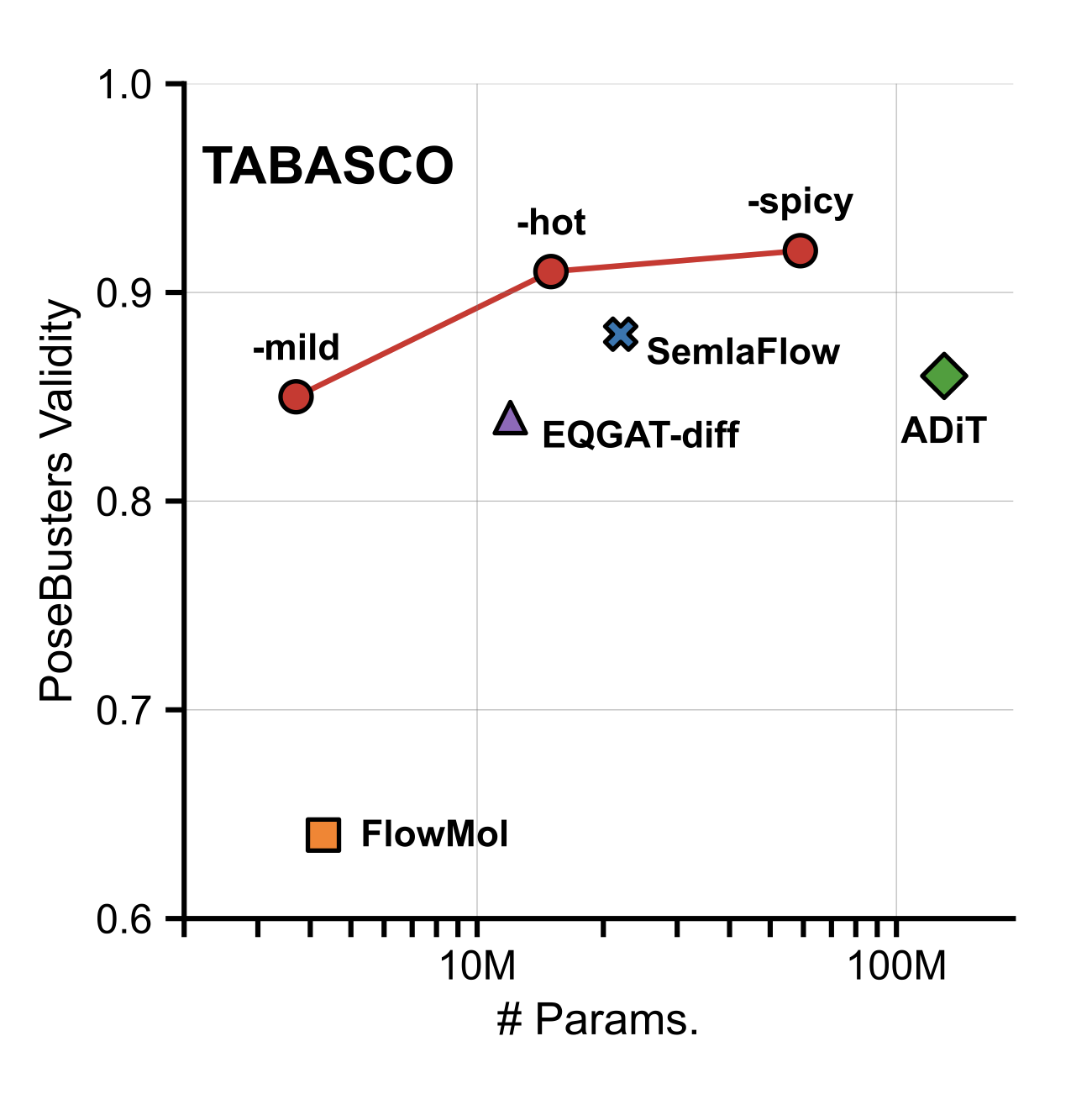
TABASCO: A Fast, Simplified Model for Molecular Generation with Improved Physical Quality
Carlos Vonessen*, Charles Harris*, Miruna Cretu*, Pietro Liò (* equal contribution)
ICML GenBio Workshop 2025
We introduce TABASCO which re- laxes these assumptions: The model has a standard non-equivariant transformer architecture, treats atoms in a molecule as sequences and re- constructs bonds deterministically after genera- tion. The absence of equivariant layers and message passing allows us to significantly simplify the model architecture and scale data through- put. On the GEOM-Drugs benchmark TABASCO achieves state-of-the-art PoseBusters validity and delivers inference roughly 10× faster than the strongest baseline, while exhibiting emergent ro- tational equivariance despite symmetry not be- ing hard-coded.

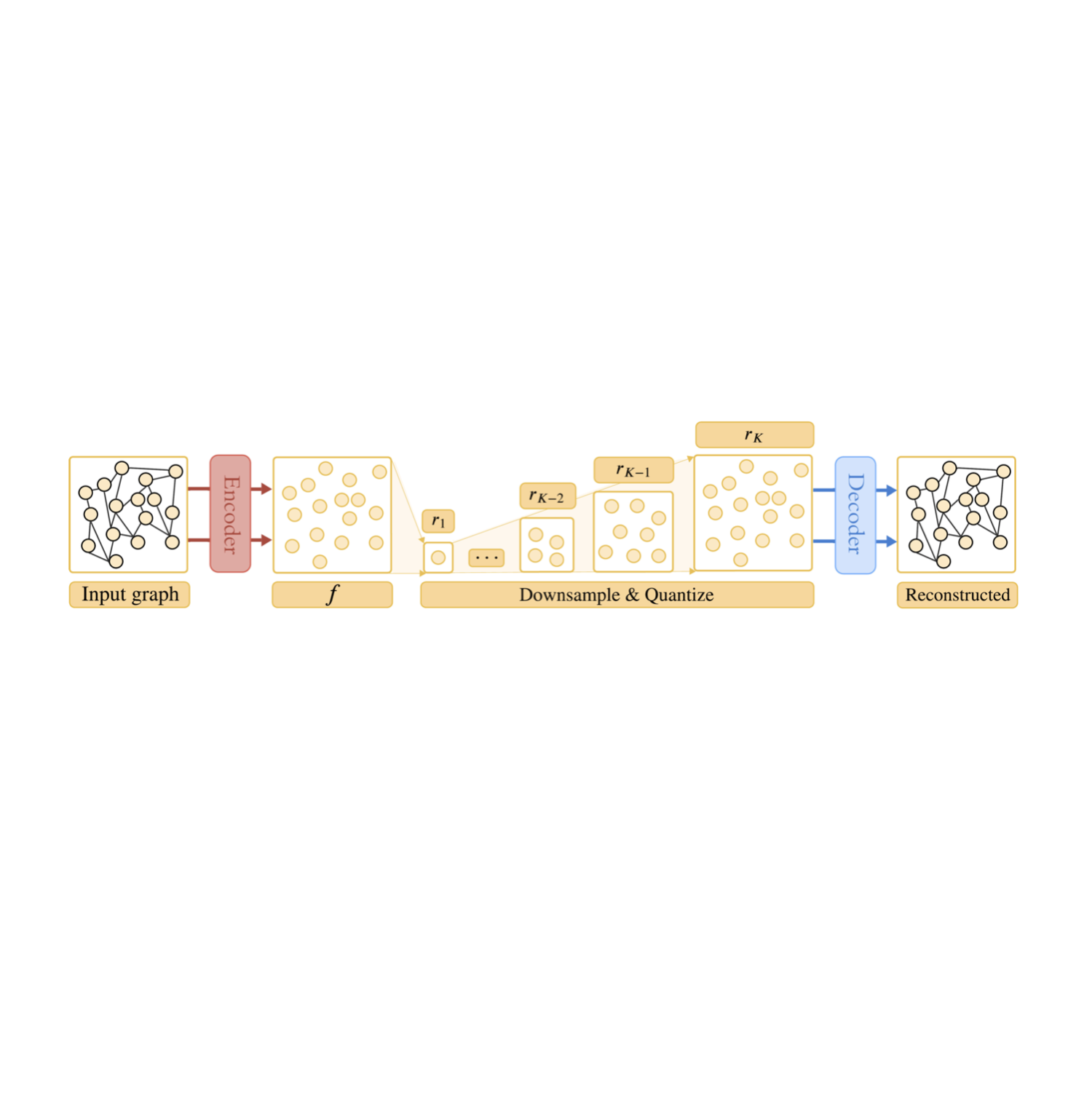
Diffusion-Free Graph Generation with Next-Scale Prediction
Samuel Belkadi, Steve Hong, Marian Chen, Miruna Cretu, Charles Harris, Pietro Liò
ICML GenBio Workshop 2025
Here, we propose MAG, a novel diffusion-free graph generation framework based on next-scale prediction. By leveraging a hierarchy of latent representations, the model proogressively generates scales of the entire graph without the need for explicit node ordering.

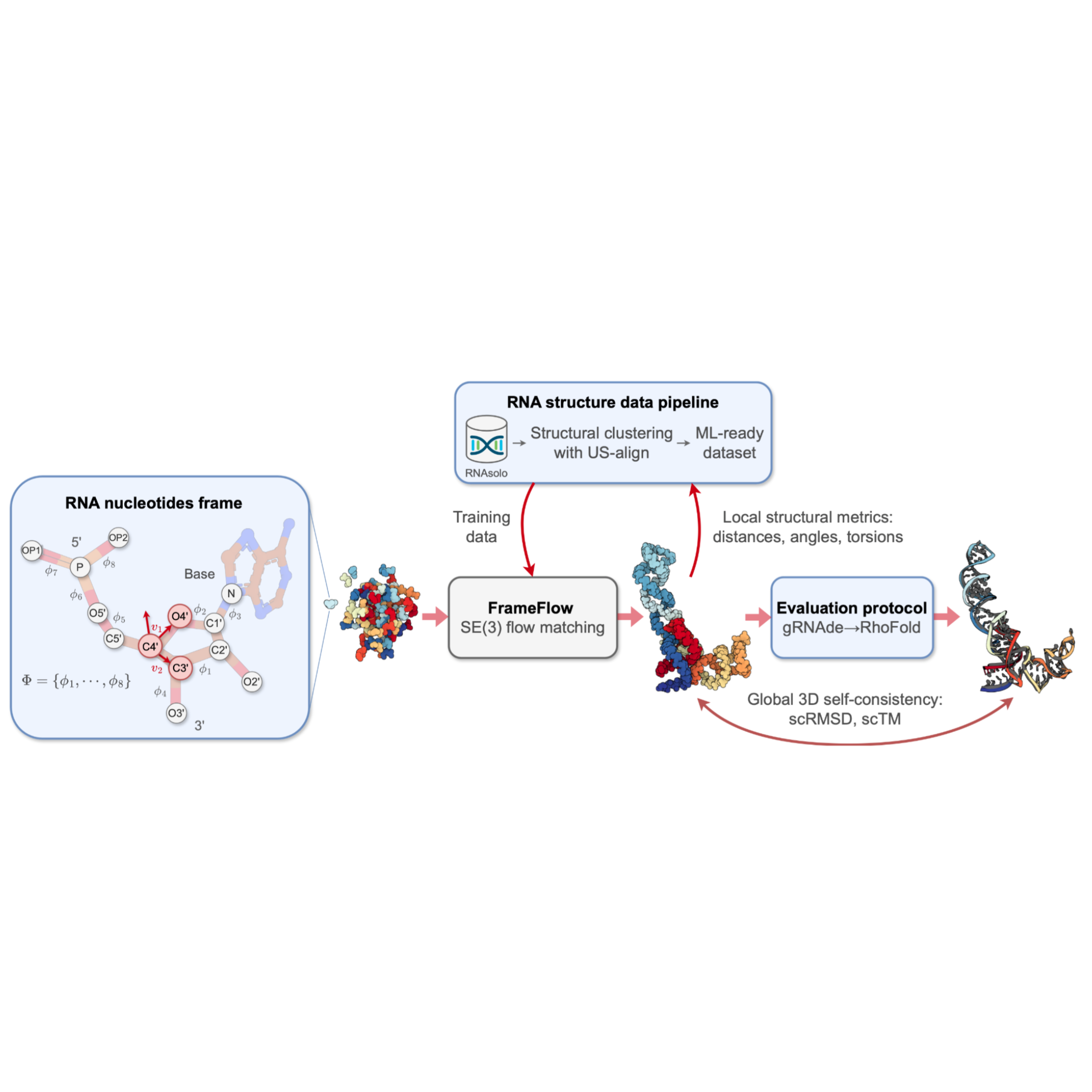
RNA-FrameFlow: Flow Matching for de novo 3D RNA Backbone Design
Rishabh Anand, Chaitanya Joshi, Alex Morehead, Arian R. Jamasb, Charles Harris, Simon Mathis, Keiran Didi, Bryan Hooi, Pietro Liò
Transactions on Machine Learning Research (TMLR) 2025 Spotlight
We introduce RNA-FrameFlow, the first generative model for 3D RNA backbone design. We build upon $SE(3)$ flow matching for protein backbone generation and establish protocols for data preparation and evaluation to address unique challenges posed by RNA modeling.

2024

DiffSBDD: Structure-based Drug Design with Equivariant Diffusion Models
Arne Schneuing*, Charles Harris*, Yuanqi Du*, Arian Jamasb, Ilia Igashov, Weitao Du, Tom Blundell, Pietro Lió, Carla Gomes, Max Welling, Michael Bronstein, Bruno Correia (* equal contribution)
Nature Computational Science 2024
DiffSBDD was one of the first equivariant diffusion models for structure-based drug design.

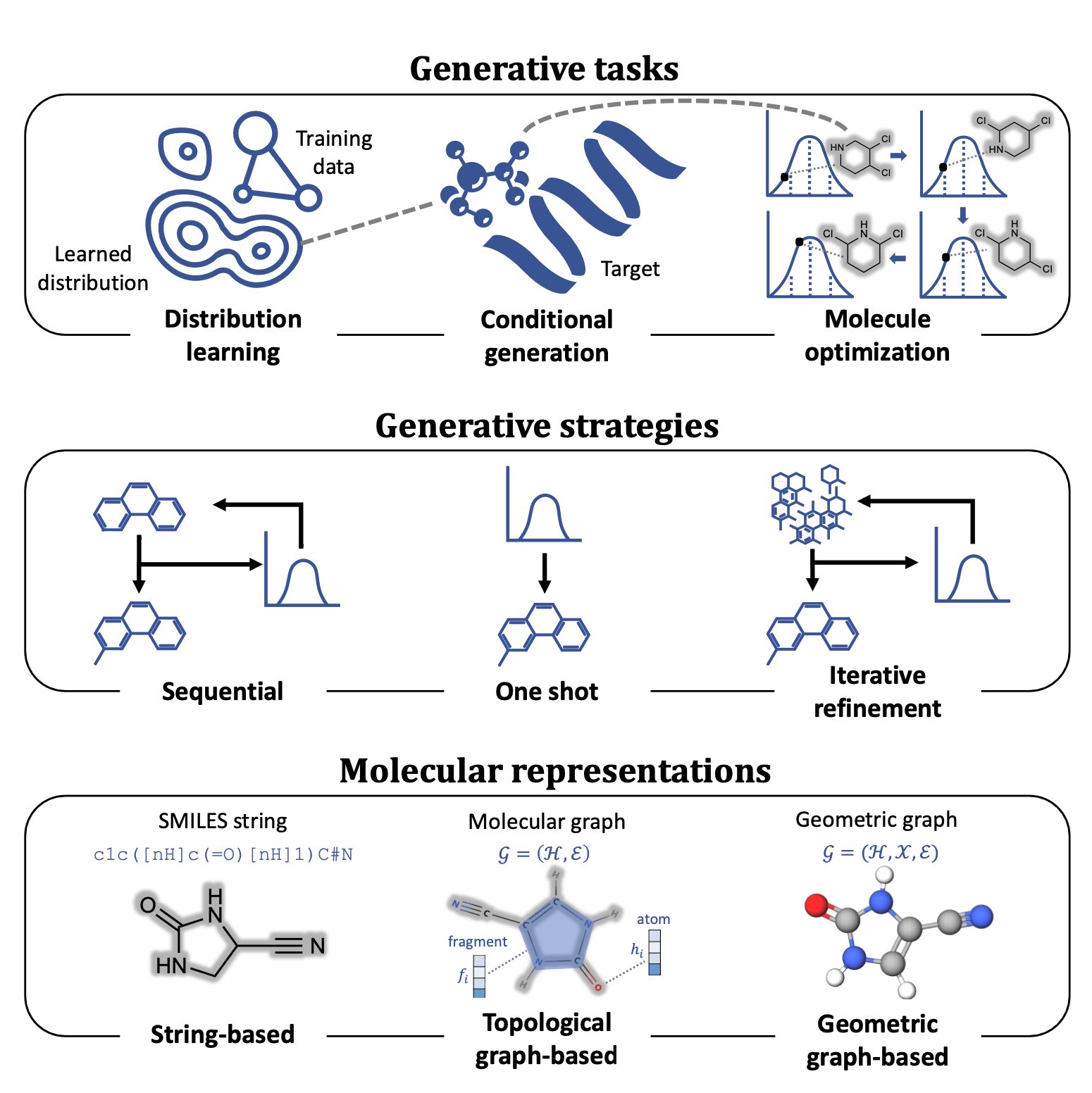
Machine learning-aided generative molecular design
Yuanqi Du, Arian R. Jamasb, Jeff Guo, Tianfan Fu, Charles Harris, Yingheng Wang, Chenru Duan, Pietro Liò, Philippe Schwaller, Tom L. Blundell
Nature Machine Intelligence 2024
Here we provide a comprehensive overview of the current state of the art in molecular design using machine learning models as well as important design decisions, such as the choice of molecular representations, generative methods and optimization strategies. Subsequently, we present a collection of practical applications in which the reviewed methodologies have been experimentally validated, encompassing both academic and industrial efforts. Finally, we draw attention to the theoretical, computational and empirical challenges in deploying generative machine learning and highlight future opportunities to better align such approaches to achieve realistic drug discovery end points.

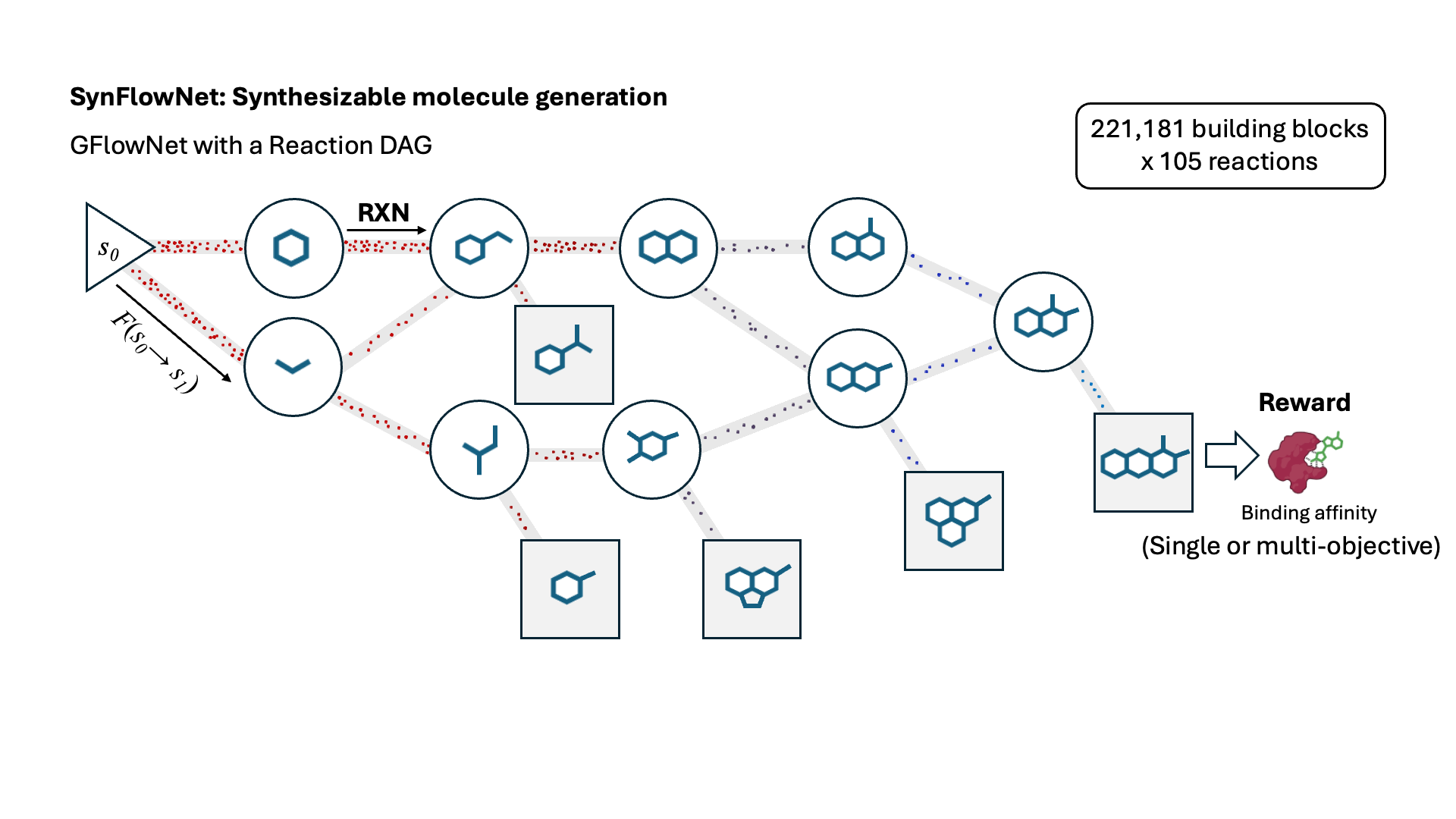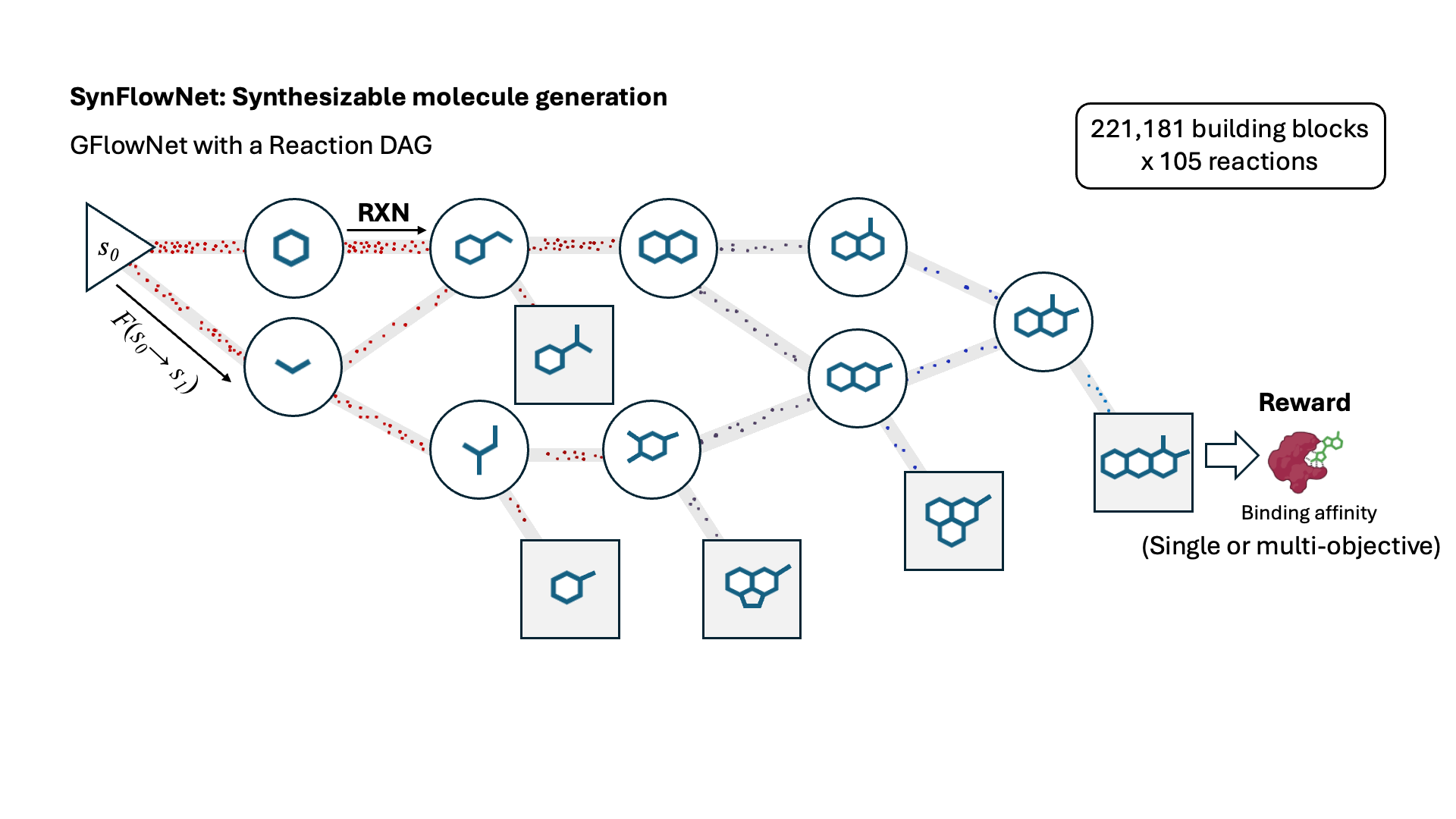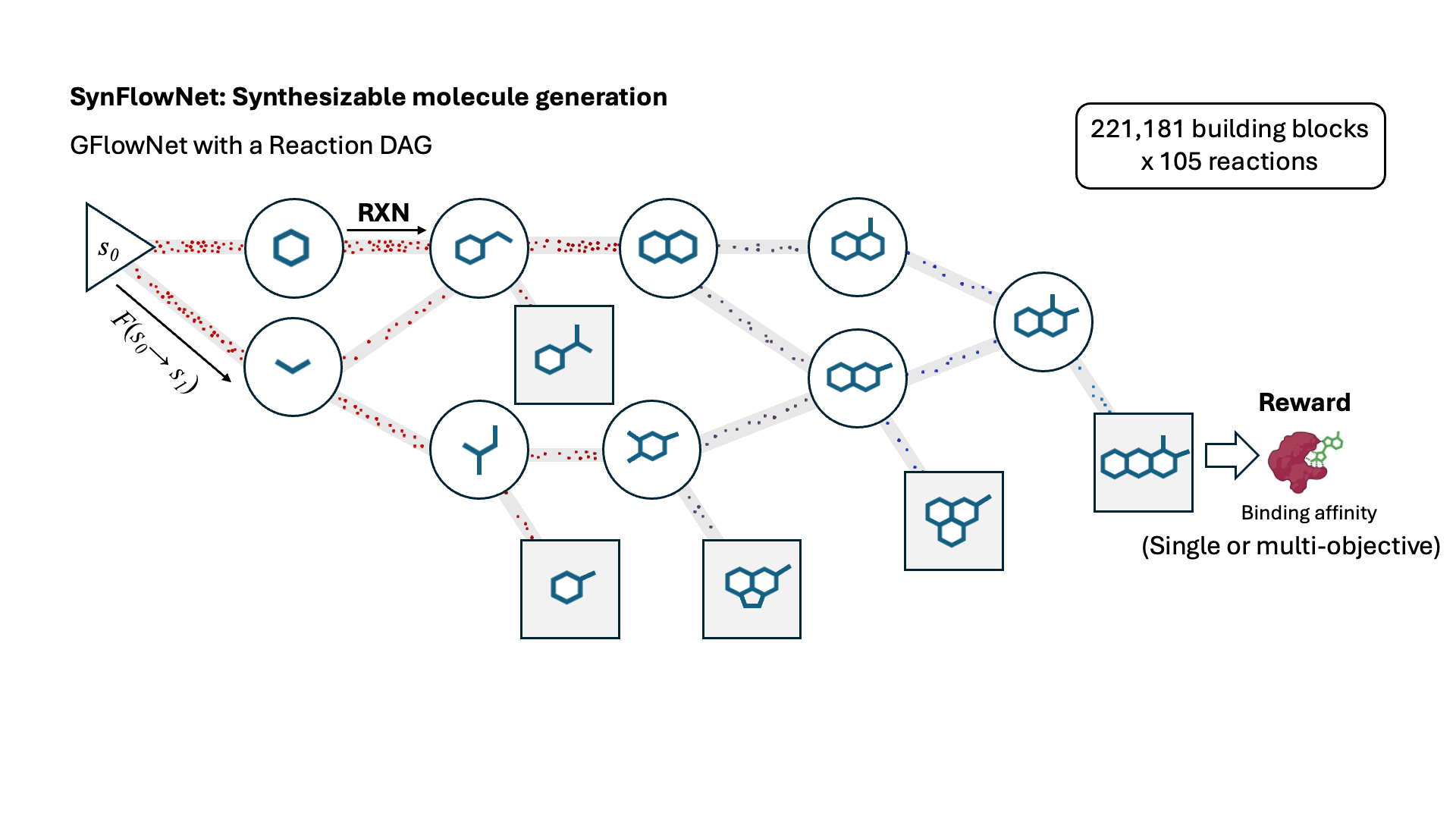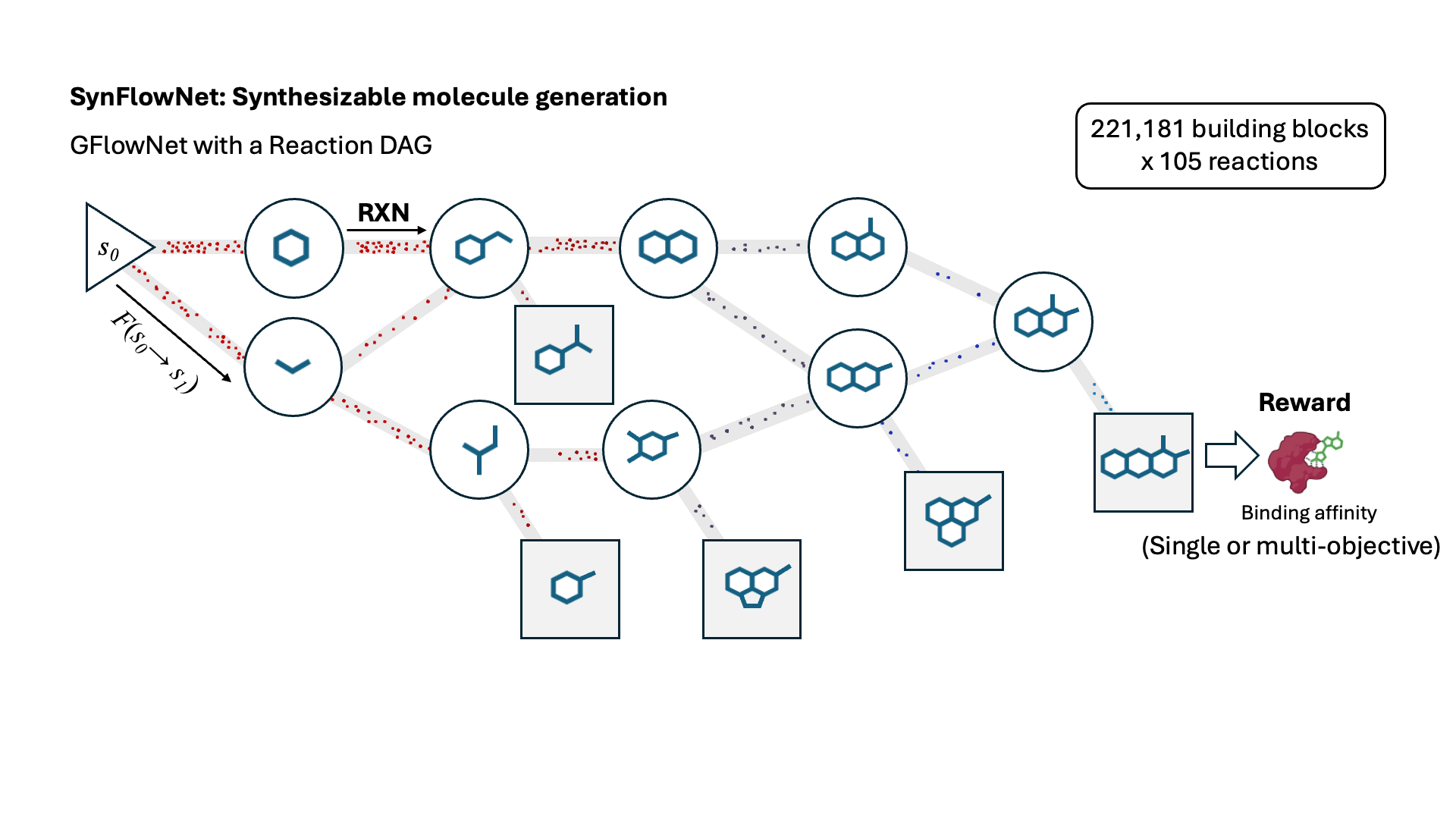
SynFlowNet: Design of Diverse and Novel Molecules with Synthesis Constraints
Miruna Cretu, Charles Harris, Ilia Igashov, Arne Schneuing, Marwin Segler, Bruno Correia, Julien Roy, Emmanuel Bengio, Pietro Liò
ICLR 2025 Spotlight
This work introduces SynFlowNet, a GFlowNet model whose action space uses chemically validated reactions and reactants to sequentially build new molecules. We evaluate our approach using synthetic accessibility scores and an independent retrosynthesis tool. SynFlowNet consistently samples synthetically feasible molecules, while still being able to find diverse and high-utility candidates.

2023
Evaluating Representation Learning on the Protein Structure Universe
Arian Jamasb, Alex Morehead, Chaitanya Joshi, Zuobai Zhang, Kieran Didi, Simon Mathis, Charles Harris, Tian Tang, Jianlin Cheng, Pietro Lió, Tom Blundell
ICLR 2024
In the benchmark, we implement numerous featurisation schemes, datasets for self-supervised pre-training and downstream evaluation, pre-training tasks, and auxiliary tasks. The benchmark can be used as a working template for a protein representation learning research project, a library of drop-in components for use in your projects, or as a CLI tool for quickly running protein representation learning evaluation and pre-training configurations.

PoseCheck: Generative Models for 3D Structure-based Drug Design Produce Unrealistic Poses
Charles Harris, Kieran Didi, Arian Jamasb, Chaitanya Joshi, Simon Mathis, Pietro Liò, Tom Blundell
MLSB Workshop @ NeurIPS 2023 Spotlight
This work introduced PoseCheck, an extensive analysis of multiple state-of-the-art methods and find that generated molecules have significantly more physical violations and fewer key interactions compared to baselines, calling into question the implicit assumption that providing rich 3D structure information improves molecule complementarity. We make recommendations for future research tackling identified failure modes and hope our benchmark will serve as a springboard for future SBDD generative modelling work to have a real-world impact.

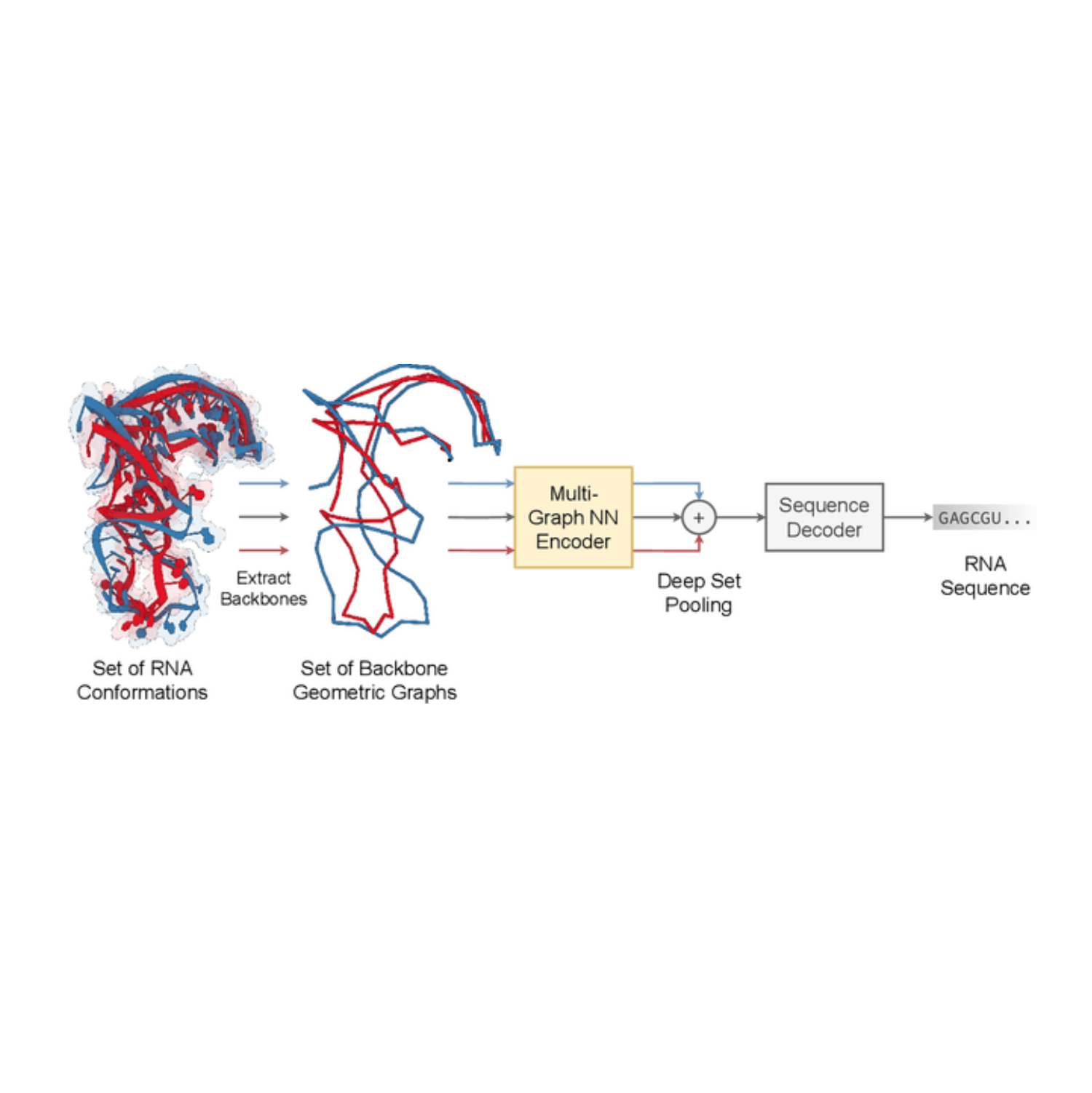
Multi-State RNA Design with Geometric Multi-Graph Neural Networks
Chaitanya Joshi, Arian Jamasb, Ramon Vinas, Charles Harris, Simon Mathis, Pietro Lió
ICLR 2025 Spotlight
This work introduces gRNAde, a geometric RNA design pipeline operating on 3D RNA backbones to design sequences that explicitly account for structure and dynamics. Under the hood, gRNAde is a multi-state Graph Neural Network that generates candidate RNA sequences conditioned on one or more 3D backbone structures where the identities of the bases are unknown.


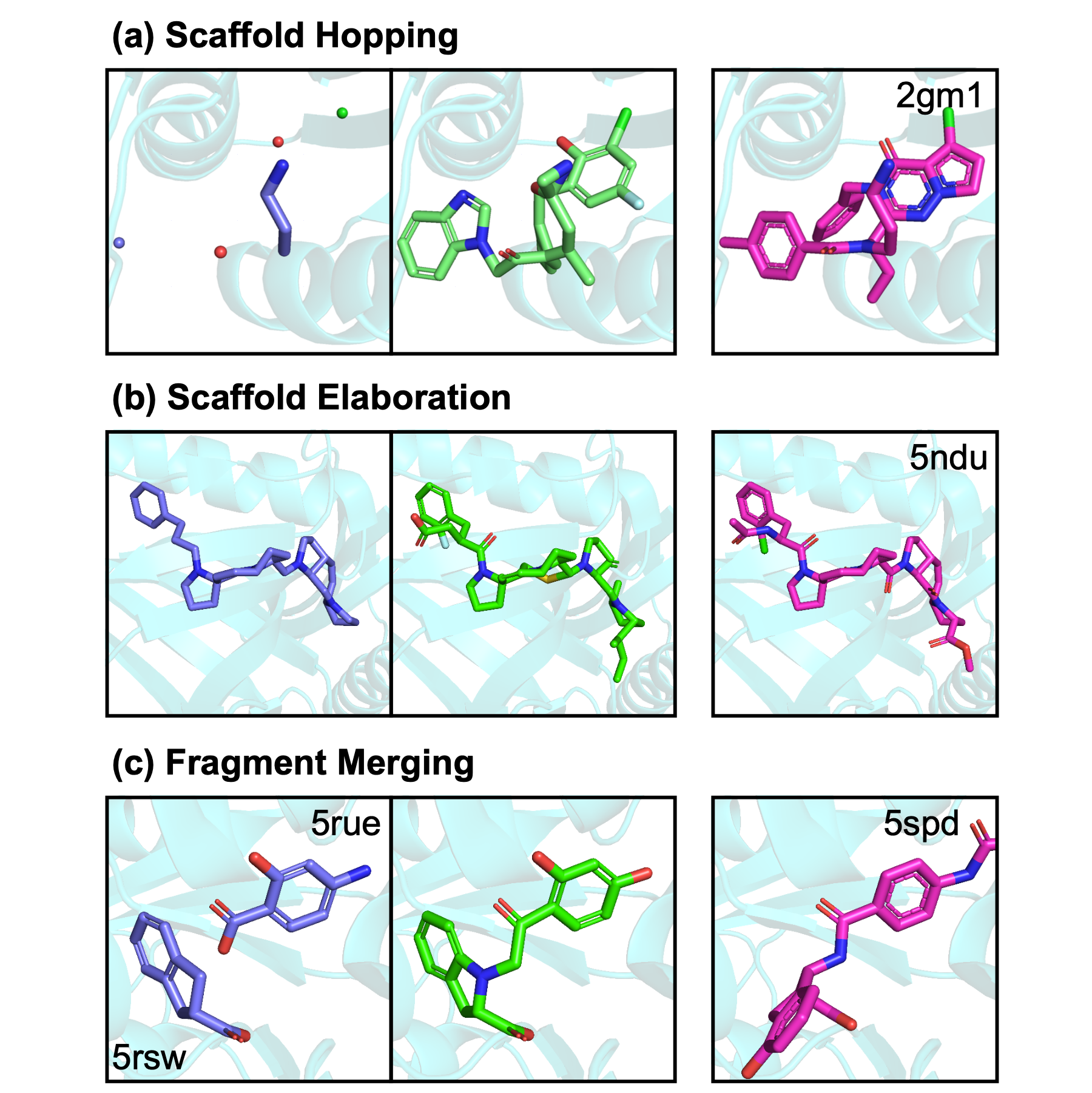
Flexible Small-Molecule Design and Optimization with Equivariant Diffusion Models
Charles Harris, Keiran Didi, Arne Schneuing, Yuanqi Du, Arian Jamasb, Michael Bronstein, Bruno Correia, Tom Blundell, Pietro Lió
MLDD @ ICLR 2023
This work extended the use of DiffSBDD to various subtasks in SBDD, such as fragment linking and compounds optimisation.

2022
Graphein - a Python Library for Geometric Deep Learning and Network Analysis on Protein Structures and Interaction Networks
Arian Jamasb, Ramon Vinas Torne, Eric Ma, Charles Harris, Ilia Igashov, Kevin Huang, Dominic Hall, Pietro Lió, Tom Blundell
NeurIPS 2022
Graphien is an easy to use package to turn structural biology data into machine learning ready data formats, notably for graph neural networks.
